Reversible processes in cell cycle control during S-phase: Dynamic ubiquitylation and deubiquitylation of the sliding clamp PCNA
The genome must be completely and faithfully replicated during S-phase before it is segregated into the progeny to ensure genomic stability. Cells are constantly exposed to the threat of endogenous and exogenous DNA damage, a major source of genomic instability and, in multicellular organisms, a driving force for cancer. Cells are particularly sensitive to damage during DNA replication because the stringent nature of replicative DNA polymerases cannot accommodate damaged bases and, consequently, DNA synthesis is blocked with potential deleterious effects. To cope with base damage during DNA replication all three domains of life have evolved damage tolerance mechanisms that allow them to circumvent DNA lesions allegedly to ensure coordinated and timely progression of replication forks. Tolerance to DNA damage is based on translesion DNA synthesis (TLS), a bypass mechanism that allows cells to overcome the potentially lethal effects of confronting DNA base lesions that impede progression of replicative DNA polymerases. DNA Damage Tolerance, or DDT, has been misleadingly known as a post-replicative repair mechanism. However, DDT is not a repair tool as DNA lesions are only circumvented as a consequence of using DNA templates containing damaged bases.
The identification of specialized, evolutionary conserved but alternative DNA polymerases with the ability to use damaged templates and effectively bypass lesions, was a landmark discovery indicating the existence of a mechanism to tolerate DNA damage. These enzymes, termed TLS DNA polymerases, are potentially mutagenic as they accommodate templates with bulky lesions, lack proofreading activity, are low-fidelity and, therefore, error-prone. An alternative mechanism of DDT exists, which is based on template switching (TS), an error-free mechanism that involves sister-strand pairing. TS was first described in prokaryotes as an error-free post-replicative repair mechanism involved in filling-in gaps left behind replication forks. TS plays a not yet fully understood role in replication all throughout the evolutive scale.
The proliferating cell nuclear antigen (PCNA) has a central role in DNA replication. Moreover, it also constitutes a central DNA scaffold for the recruitment of a plethora of components involved in both tolerance and response to DNA damage mechanisms. In eukaryotes, DDT mechanisms are mediated by ubiquitylation of this essential replication factor PCNA, a key processivity factor for polymerases during DNA synthesis. Monoubiquitylation of Lys164 of PCNA (ubPCNAK164) promotes TLS-mediated error-prone translesion synthesis. Further Lys63-linked polyubiquitylation of mono-ubPCNAK164 triggers the error-free mechanism based on TS. The prevailing view is that PCNAK164 is ubiquitylated in a DNA damage-dependent manner. Therefore, it is believed that not such modification (ubPCNAK164) occurs in cells during unchallenged S-phase in most organisms except in fission yeast and Xenopus.
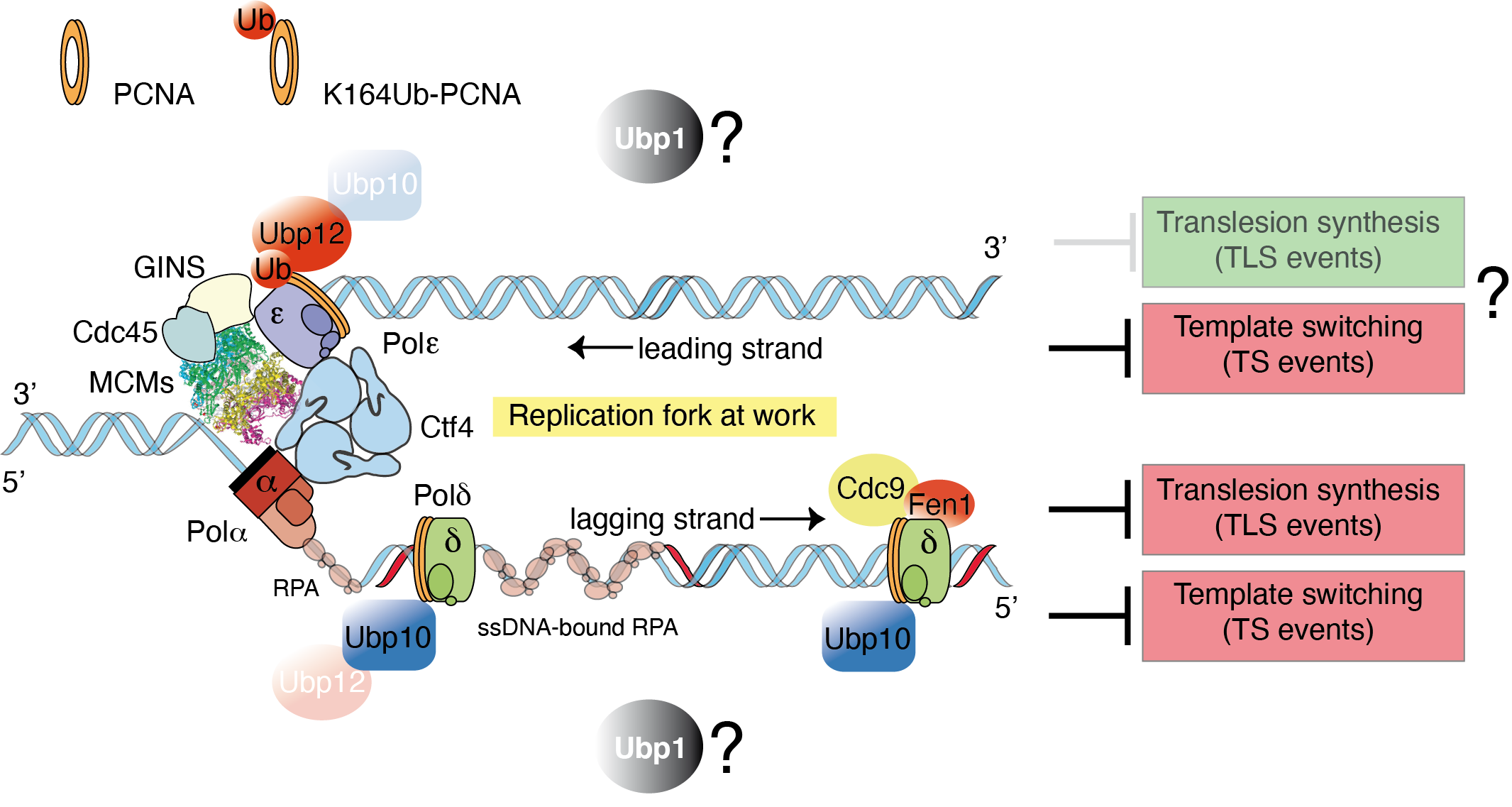
Although the evolutionary conserved mechanism of PCNA ubiquitylation is well understood, the deubiquitylation of ubPCNA remains uncharacterized. Our group is interested in understanding the role and, therefore, the biological significance of ubPCNA deubiquitylation in yeast models. Our working hypothesis is that Ubiquitin-specific proteases, or PCNA-DUBs, revert PCNA ubiquitylation to prevent excessive translesion synthesis (TLS) on replicating chromatin. This hypothesis predicts that both branches of the DNA damage tolerance pathway, TLS-DNA polymerases sampling on replicating DNA and template switching during S-phase, would be limited at forks by PCNA deubiquitylation.
Research Aims
Given the tremendous impact that PCNA-DUBs ubiquitin-proteases may have in genomic stability, the major aim of our research group is to understand in depth the replication fork-coupled mechanism down-regulating DDT events through PCNAdeubiquitylation.
Despite recent progress in the DNA damage tolerance field, a number of important questions remain open. We want to address what we consider the most appealing questions in the field to gain a clear understanding of what is in full the role of PCNA-DUBs at sites of nascent DNA. Particularly, we want to address the following questions:
a) Is the control of DDT at replication forks asymmetric and, consequently, divergent in leading and lagging strands?
b) Is leading strand DNA synthesis at forks permissive for the error-prone branch of DDT?
c) What is the molecular nature of the template switching events at forks prevented by PCNA-DUBs?
d) Does fork-coupled PCNA-DUBs integrate Okazaki-fragments maturation with reversion of PCNA ubiquitylation?
Goals Achieved
- Smooth and timely progression through S-phase in eukaryotic cells requires PCNAK164 deubiquitylation.
- Ubp10 and Ubp12 budding yeast ubiquitin proteases remove ubiquitin from PCNAK164 with discrete chain preferences.
- Replication fork association of Ubp10, but not Ubp12, depends on PCNA binding to lagging strands.
- PCNA-DUBs Ubp10 and Ubp12 counteract template-switching events and Rev1, a key translesion synthesis adaptor, binding to replication forks.
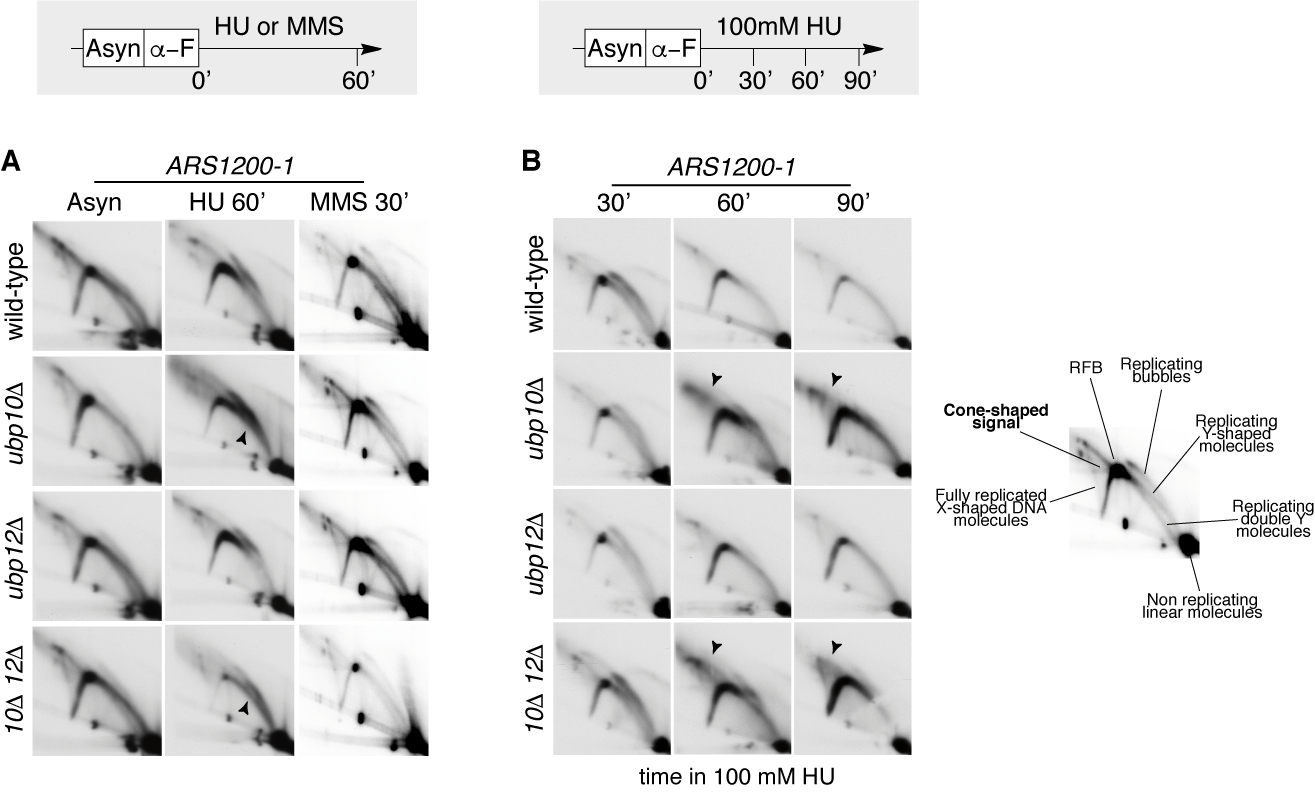
PCNA-DUB mutants ubp10 ubp12 accumulate non-canonical small-Y molecules and cone-shaped signals at HU-stalled forks. (A) Wild-type cells and ubp10, ubp12 and ubp10 ubp12 mutant cells were synchronized in G1 and released in the presence of 200mM HU or 0.033% MMS. Samples were taken after 60 minutes and processed for 2D-gel analysis. Membranes were hybridized to a probe spanning ARS1200-1 replication origin. (B) Wild-type cells and ubp10, ubp12 and ubp10 ubp12 mutant cells were synchronized in G1 and released in the presence of 100mM HU. Samples were taken at indicated intervals and processed for 2D-gel analysis. Membranes were also hybridized to a probe spanning ARS1200-1 replication origin. A Schematic of canonical replication intermediates and fully replicated joint molecules detected by 2D-gel analysis at ARS1200 is shown. Canonical small-Ys (black arrow in A) reflect passive replication by forks emanating outside the probed fragment. Con-shaped non-canonical intermediates in B are pointed by and arrow.
Future Goals
- Analysis of additional ubiquitin-proteases involved in PCNA deubiquitylation.
- Testing asymmetry in DNA Damage Tolerance regulation at replication forks.
- Analysis of non-canonical small Y-shaped replication intermediates accumulating at stalled forks in ubp10 and ubp10 ubp12mutants.
Specific Objetives
1. Testing interactions in vivo of PCNA-DUBs with replisome components.
The goal is to identify the interactome of PCNA-DUBs at active replication forks using the isolation of proteins on nascent DNA(iPOND)-method coupled with mass spectrometry. We will validate that PCNA-DUBs are Replisome components during normal and perturbed S-phase. Identifying proteins that interact with PCNA-DUBs and their posttranslational modifications is an additional key element to understanding how the genome is replicated every cell division cycle. One effective experimental approach to analyse systematically the components, as well as their modifications, of the replisome progression complex is the iPOND method. The purification and identification by mass spectrometry of proteins on newly synthesized DNA using iPOND will allow us to extend the study of the potential temporal association of PCNA-DUBs with replication forks and also to confirm that they are functional components of the replisome as we hypothesized.
2. eSPAN asymmetry test for PCNA-DUBs and TLS-polymerases.
eSPAN is a strand-specific analysis based on the enrichment and Sequencing of Protein-Associated to Nascent DNA. eSPAN measures the relative amounts of proteins at nascent leading and lagging strands of DNA replication forks in a step-wise procedure involving chromatin immunoprecipitation of a protein of interest (bait) followed by the enrichment of protein-associated nascent DNA through BrdU immunoprecipitation. The key part is that the isolated ssDNA is then subjected to strand-specific sequencing allowing detection of proteins enriched at leading or lagging strand of nascent DNA. We plan to test asymmetry of PCNA-DUBs and TLS DNA-polymerases at forks by a strand-specific analysis based on eSPAN.
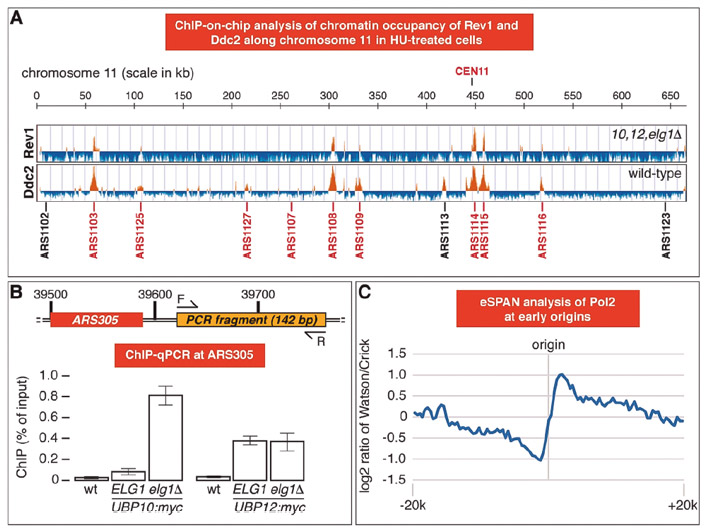
Genome-wide analysis by chromatin immunoprecipitation of key components of DNA damage tolerance pathways. (A) ChIP-on-chip analysis of REV1-myc ubp10Δ ubp12Δ elg1Δ (10, 12, elg1Δ) and REV1-myc (wild-type) cells pre-synchronized with α-factor and released for 1 hour in presence of 0.2 M HU. Orange histogram bars in the Y-axis show the average signal ratio of loci significantly enriched in each immunoprecipitated fraction in log2 scale. The X-axis shows chromosomal coordinated in kilobases along regions in chromosome XI Early-firing (in red) and late/dormant (in black) replication origins are indicated. (B) Deletion of ELG1 influences Ubp10, but not Ubp12, association to ARS305. The indicated cells were cultured as in (A) and processed for ChIP-qPCR. Means and standard deviations of three independent experiments are shown. A schematic of the ARS305-proximal DNA fragment amplified is provided. (C) Strand-specific test analysis of the catalytic subunit of DNA polymerase epsilon (POL2) made by Rodrigo Bermejo’s group (CIB Margarita Salas) in collaboration with us.
We are currently working on the following research projects:
We are focused to understanding how processive DNA synthesis is ensured in the presence of damaged templates and whether PCNA-DUBs asymmetric localisation at replication forks have any functional implications for DDT control.
I. Analysis of PCNAK164 ubiquitin-proteases (PCNA-DUBs) as regulatory replisome components.
II. Testing asymmetry in DNA damage tolerance regulation at replication forks.
III. Studying the role of PCNA deubiquitylation in Okazaki-fragments maturation